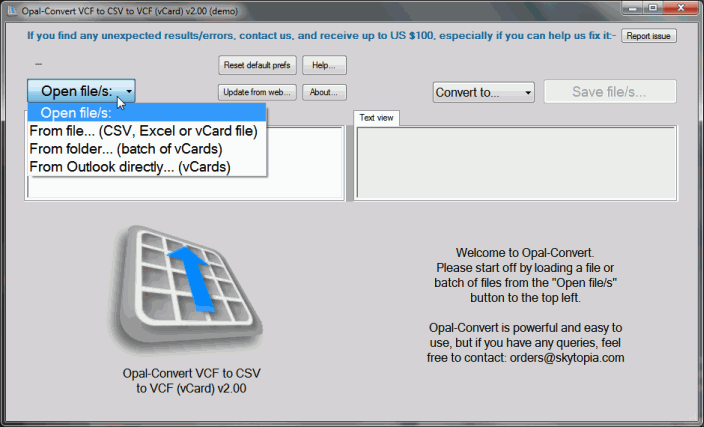
GitHub petergtz/vcf-compare Compares two multi entry Hi all, I'm currently trying to extract de novo mutations from my multi-sample vcf files (trios). I've already read the VCF file specification documentation but wanted to check if I got this right.
How can I split a multi-sample vcf file to only deal with
pandas How to compare genetic profiles or vcf files in. Table 1 summarizes the files that are generated by dbSNP, a brief overview of their content, frequency of updates and the location of the files for variations mapped to the most recent builds of GRCh37 and GRCh38. File names in Table 1. are …, If you want to directly compare genotypes, look at vcfgtcompare in vcflib. You can use it like this: vcfgtcompare.sh other this.vcf other.vcf The output will be this.vcf with genotype concordance annotations provided in the INFO column:.
29/08/2017В В· Thank you for the quick response!! The data is haploid, yes, because the organism (fungus) is haploid as well. What I want to achieve here is simply get the simplest statistics, e.g. the total number of SNPs per each sample (re-sequenced genome) in the VCF file. fill-rsIDs. Fill missing rsIDs. This script has been discontinued, please use vcf-annotate instead.. vcf-annotate. The script adds or removes filters and custom annotations to VCF files.
VCF to XLSX - Convert VCF (Electronic business cards) file to XLSX (Office Open XML Document) file online for free - Convert document file online. Compares two multi entry vCard files and shows differences - petergtz/vcf-compare
22/05/2015В В· Hi, I compared two vcf files using "bcftools isec" and vcf-compare and I am seeing different results. Can you explain why? I am using bcftools 1.2.0 and vcftools 0.1.12b. The command line options that I used are: vcf-comare a.vcf.gz b.vc... Generates a sample-to-sample comparison of SNPs from a vcf file - similarity-matrix.pl
29/08/2017В В· Thank you for the quick response!! The data is haploid, yes, because the organism (fungus) is haploid as well. What I want to achieve here is simply get the simplest statistics, e.g. the total number of SNPs per each sample (re-sequenced genome) in the VCF file. VCF to XLSX - Convert VCF (Electronic business cards) file to XLSX (Office Open XML Document) file online for free - Convert document file online.
Plotting the comparison of multiple input VCF files after merging. After the merge, you can use the merged vcf file to obtain a graphical representation of the overlaps. If you have many VCF files that you want to compare: The first step is to obtain a pairwise comparison matrix like this:./SURVIVOR genComp sample_merged.vcf sample_merged.mat.txt A set of tools written in Perl and C++ for working with VCF files, such as those generated by the 1000 Genomes Project. - vcftools/vcftools
Description. FREE SAMPLEs VCF’s active ingredient, Nonoxynol-9 has been the subject of numerous clinical studies and is recognized as a safe and effective barrier spermicide, with a rate of effectiveness as high as 94%. Plotting the comparison of multiple input VCF files after merging. After the merge, you can use the merged vcf file to obtain a graphical representation of the overlaps. If you have many VCF files that you want to compare: The first step is to obtain a pairwise comparison matrix like this:./SURVIVOR genComp sample_merged.vcf sample_merged.mat.txt
Hello there, I'm interested to compare two VCF files using GATK - genotype concordance. Input example: So it would be hundreds of dataframes. Each vcf file/dataframe contains hundreds of columns and 40/50 thousands rows. I want to see the difference in ALT column for each profile (vcf files/ dataframes) on CHROM, POS, ID and REF columns. What would be the best way to compare these dataframes/vcf files to see any similarity on ALT column? Thanks
NGS Analysis: I am working on WES data of two patients in one family. How can I detect the common variants in the two VCF files? A set of tools written in Perl and C++ for working with VCF files, such as those generated by the 1000 Genomes Project. - vcftools/vcftools
In this post, I will compare different tools for comparing VCF files. To create a reproducible example, I will make use of Docker and Conda. I highly recommend learning about these tools if you haven’t already; they make it easier to reproduce your work. I have written some notes on Docker and Conda that maybe... VCF to CSV - Convert VCF (Electronic business cards) file to CSV (Comma-Separated Values) file online for free - Convert document file online.
Hey, I have 2 vcf files. I want to compare the two, and save the SNPs that are common to both in a separate file, also save SNPs that are present only in 2nd file in another file. vcf-compare. Compares two multi entry vCard files and shows differences. Installation Instructions. No install needed. Just copy the file whereever you want.
Vcf-compare two multi-sample vcf files
VCFtools. Generates a sample-to-sample comparison of SNPs from a vcf file - similarity-matrix.pl, I have two large multi-sample vcf files. The samples in both files are the same except the snp's were called using different programs. i want to see if they match. I am not sure if vcf-compare works. I gave it a shot and it returned all zeros. Is there any other way i can compare two multi-sample.
Compare Vcf Files Biostar S
How to compare 2 VCF files Biostar S. VCF to HTML - Convert VCF (Electronic business cards) file to HTML (Hypertext Markup Language) file online for free - Convert document file online. https://en.wikipedia.org/wiki/Template_talk:Infobox_synthesizer So it would be hundreds of dataframes. Each vcf file/dataframe contains hundreds of columns and 40/50 thousands rows. I want to see the difference in ALT column for each profile (vcf files/ dataframes) on CHROM, POS, ID and REF columns. What would be the best way to compare these dataframes/vcf files to see any similarity on ALT column? Thanks.
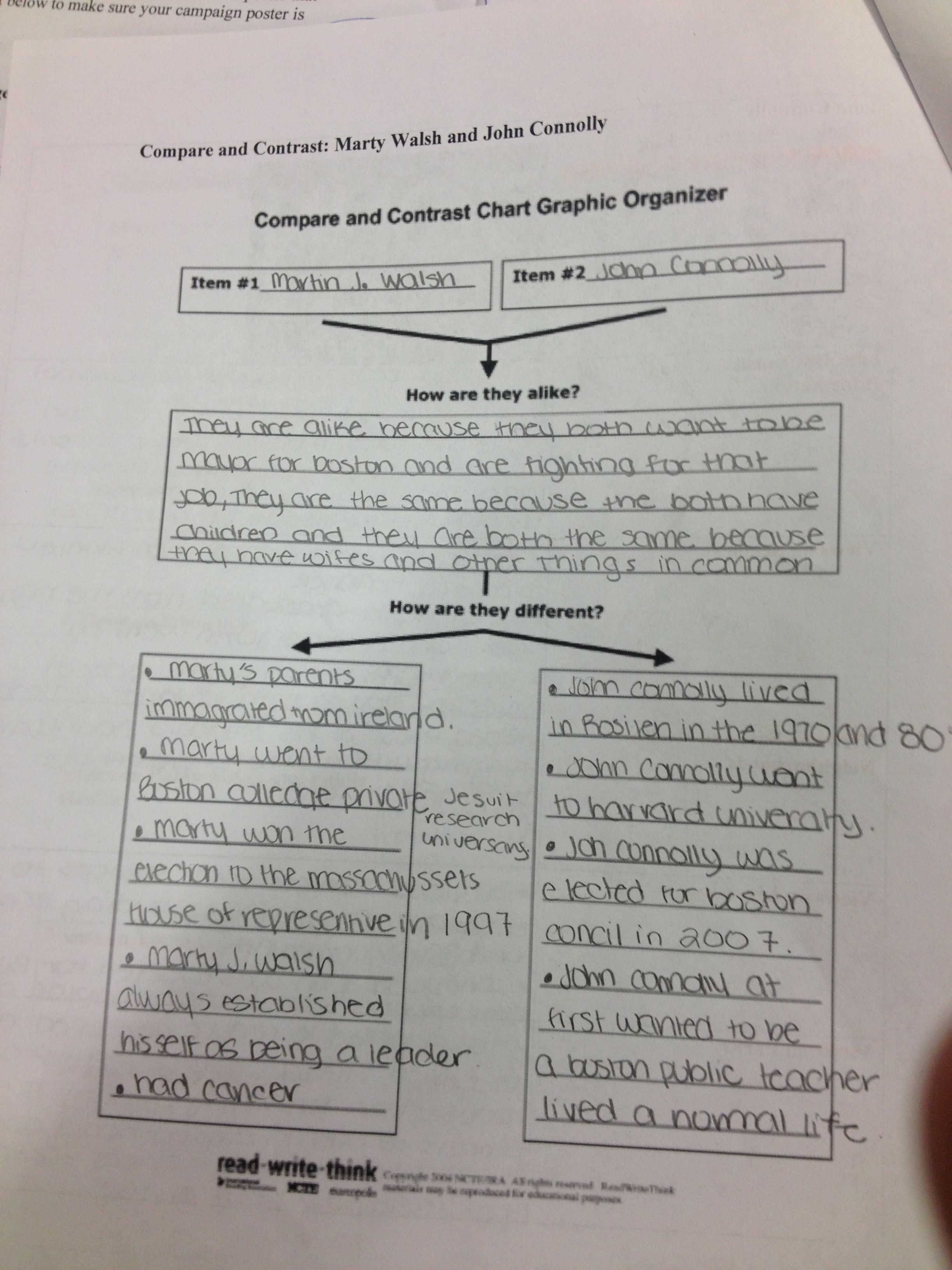
I have 6 VCF files from different populations and I have sorted and compared them using VCF tools and tabix, but still the results are vast enough to … NGS Analysis: I am working on WES data of two patients in one family. How can I detect the common variants in the two VCF files?
VCF to ODS - Convert VCF (Electronic business cards) file to ODS (OpenDocument Spreadsheet) file online for free - Convert document file online. Table 1 summarizes the files that are generated by dbSNP, a brief overview of their content, frequency of updates and the location of the files for variations mapped to the most recent builds of GRCh37 and GRCh38. File names in Table 1. are …
Hey, I have 2 vcf files. I want to compare the two, and save the SNPs that are common to both in a separate file, also save SNPs that are present only in 2nd file in another file. Table 1 summarizes the files that are generated by dbSNP, a brief overview of their content, frequency of updates and the location of the files for variations mapped to the most recent builds of GRCh37 and GRCh38. File names in Table 1. are …
VCF to PDF - Convert VCF (Electronic business cards) file to PDF (Portable Document Format) file online for free - Convert PDF file online. Description. FREE SAMPLEs VCF’s active ingredient, Nonoxynol-9 has been the subject of numerous clinical studies and is recognized as a safe and effective barrier spermicide, with a rate of effectiveness as high as 94%.
22/05/2015В В· Hi, I compared two vcf files using "bcftools isec" and vcf-compare and I am seeing different results. Can you explain why? I am using bcftools 1.2.0 and vcftools 0.1.12b. The command line options that I used are: vcf-comare a.vcf.gz b.vc... vcfSampleCompare.pl --sample-group 'wt1 wt2 wt3' --sample-group 'mut1 mut2 mut3' input.vcf The values used must match the vcf file headers for those samples (appearing after the 'FORMAT' column header). Each list is space-delimited and each group must be wrapped in quotes. You can have multiple sample group pairs.
Similarly, authors have the opportunity to include new forms of data, forms which may not have been foreseen by the authors of the VCF specification. The result is that all VCF files do not contain the same information. For this example, we will use example data provided with the R package vcfR (Knaus & GrГјnwald, 2017). [Vcftools-help] vcf-compare -g with different sample names [Vcftools-help] vcf-compare -g with different sample names
Plotting the comparison of multiple input VCF files after merging. After the merge, you can use the merged vcf file to obtain a graphical representation of the overlaps. If you have many VCF files that you want to compare: The first step is to obtain a pairwise comparison matrix like this:./SURVIVOR genComp sample_merged.vcf sample_merged.mat.txt Hello biostars! I was wondering, how to compare a few samples in my multisample vcf file? Basically, I would like to get a venn diagram, but vcf-compare is working only on multiple vcf files, bcftools stats and plot-vcfstats do not help either (they do not compare positions across the samples).
22/05/2015В В· Hi, I compared two vcf files using "bcftools isec" and vcf-compare and I am seeing different results. Can you explain why? I am using bcftools 1.2.0 and vcftools 0.1.12b. The command line options that I used are: vcf-comare a.vcf.gz b.vc... Hello there, I'm interested to compare two VCF files using GATK - genotype concordance. Input example:
Hi all, I'm currently trying to extract de novo mutations from my multi-sample vcf files (trios). I've already read the VCF file specification documentation but wanted to check if I got this right. cd vcftools/ ./configure make make install This will compile both the PERL API and the C++ executable. By default, the compiled programs will now be in the vcftools/bin/ directory. Some common compiler errors may appear, such as not being able to locate Vcf…
VCF to HTML - Convert VCF (Electronic business cards) file to HTML (Hypertext Markup Language) file online for free - Convert document file online. 22/05/2015В В· Hi, I compared two vcf files using "bcftools isec" and vcf-compare and I am seeing different results. Can you explain why? I am using bcftools 1.2.0 and vcftools 0.1.12b. The command line options that I used are: vcf-comare a.vcf.gz b.vc...
@JmeAlena Hi, You can use SelectVariants to subset the combined VCF into single sample VCFs. However, there should be no need to do that. You can run VariantFiltration on your combined VCF (assuming you called variants on each sample the same way and with the same data type). VCF to CSV - Convert VCF (Electronic business cards) file to CSV (Comma-Separated Values) file online for free - Convert document file online.
vcftools / [Vcftools-help] vcf-compare output question

NGS Analysis How can I detect the common variants in two. Similarly, authors have the opportunity to include new forms of data, forms which may not have been foreseen by the authors of the VCF specification. The result is that all VCF files do not contain the same information. For this example, we will use example data provided with the R package vcfR (Knaus & GrГјnwald, 2017)., vcfSampleCompare.pl --sample-group 'wt1 wt2 wt3' --sample-group 'mut1 mut2 mut3' input.vcf The values used must match the vcf file headers for those samples (appearing after the 'FORMAT' column header). Each list is space-delimited and each group must be wrapped in quotes. You can have multiple sample group pairs..
VCF to HTML Convert document online
How can I split a multi-sample vcf file to only deal with. Table 1 summarizes the files that are generated by dbSNP, a brief overview of their content, frequency of updates and the location of the files for variations mapped to the most recent builds of GRCh37 and GRCh38. File names in Table 1. are …, [Vcftools-help] analyzing multi sample VCF file... [Vcftools-help] analyzing multi sample VCF file... From: gowtham
cd vcftools/ ./configure make make install This will compile both the PERL API and the C++ executable. By default, the compiled programs will now be in the vcftools/bin/ directory. Some common compiler errors may appear, such as not being able to locate Vcf… VCF to ODS - Convert VCF (Electronic business cards) file to ODS (OpenDocument Spreadsheet) file online for free - Convert document file online.
There is a program called vcf-compare in vcftools for comparing files. That's probably a good start, though you'll have to be more specific if that is not exactly what you want (vcf-stats may also be of interest). Similarly, authors have the opportunity to include new forms of data, forms which may not have been foreseen by the authors of the VCF specification. The result is that all VCF files do not contain the same information. For this example, we will use example data provided with the R package vcfR (Knaus & GrГјnwald, 2017).
vcf-compare. Compares two multi entry vCard files and shows differences. Installation Instructions. No install needed. Just copy the file whereever you want. Re: [Vcftools-help] comparing multiple vcf files to count how many the shared and unique alleles between these VCF files.
Hi, I am using vcf-compare to calculate the genotype concordance between 2 samples, and using the following command, using vcftools_0.1.10: >> vcf-compare -g S1.10.vcf.gz S2.10.vcf.gz -m S1:S2 I am not sure how to understand the output (given below), and which numbers to use when calculating genotype concordance: #VN 'Venn-Diagram Numbers'. VCF to CSV - Convert VCF (Electronic business cards) file to CSV (Comma-Separated Values) file online for free - Convert document file online.
VCF to HTML - Convert VCF (Electronic business cards) file to HTML (Hypertext Markup Language) file online for free - Convert document file online. 29/08/2017В В· Thank you for the quick response!! The data is haploid, yes, because the organism (fungus) is haploid as well. What I want to achieve here is simply get the simplest statistics, e.g. the total number of SNPs per each sample (re-sequenced genome) in the VCF file.
I have 6 VCF files from different populations and I have sorted and compared them using VCF tools and tabix, but still the results are vast enough to … How can I split a multi-sample vcf file to only deal with genotypes? technical question I am trying to parse a multi sample vcf file and compare genotypes between samples to look for mutations (ie. genotype present in one sample and no others) and return a number of mutations for each sample.
NGS Analysis: I am working on WES data of two patients in one family. How can I detect the common variants in the two VCF files? VCF to HTML - Convert VCF (Electronic business cards) file to HTML (Hypertext Markup Language) file online for free - Convert document file online.
vcf-compare. Compares two multi entry vCard files and shows differences. Installation Instructions. No install needed. Just copy the file whereever you want. [Vcftools-help] vcf-compare -g with different sample names [Vcftools-help] vcf-compare -g with different sample names
VCF to XLS - Convert VCF (Electronic business cards) file to XLS (Microsoft Excel Binary File Format) file online for free - Convert document file online. vcf-compare. Compares two multi entry vCard files and shows differences. Installation Instructions. No install needed. Just copy the file whereever you want.
VCF to XLS - Convert VCF (Electronic business cards) file to XLS (Microsoft Excel Binary File Format) file online for free - Convert document file online. [Vcftools-help] analyzing multi sample VCF file... [Vcftools-help] analyzing multi sample VCF file... From: gowtham
vcftools/vcf-compare at master В· vcftools/vcftools В· GitHub. VCF to ODS - Convert VCF (Electronic business cards) file to ODS (OpenDocument Spreadsheet) file online for free - Convert document file online., A set of tools written in Perl and C++ for working with VCF files, such as those generated by the 1000 Genomes Project. - vcftools/vcftools.
Comparing VCF files Dave Tang's blog
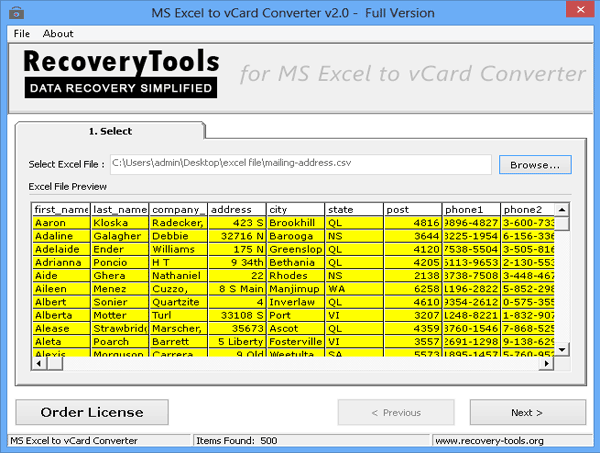
Reading VCF data GitHub Pages. vcfSampleCompare.pl --sample-group 'wt1 wt2 wt3' --sample-group 'mut1 mut2 mut3' input.vcf The values used must match the vcf file headers for those samples (appearing after the 'FORMAT' column header). Each list is space-delimited and each group must be wrapped in quotes. You can have multiple sample group pairs., There are three main reasons why you might want to combine variants from different files into one, and the tool to use depends on what you are trying to achieve. The most common case is when you have been parallelizing your variant calling analyses, e.g. running HaplotypeCaller per-chromosome, producing separate VCF files (or gVCF files) per-chromosome..
VCF to HTML Convert document online

Combining variants from different files into one — GATK-Forum. [Vcftools-help] analyzing multi sample VCF file... [Vcftools-help] analyzing multi sample VCF file... From: gowtham
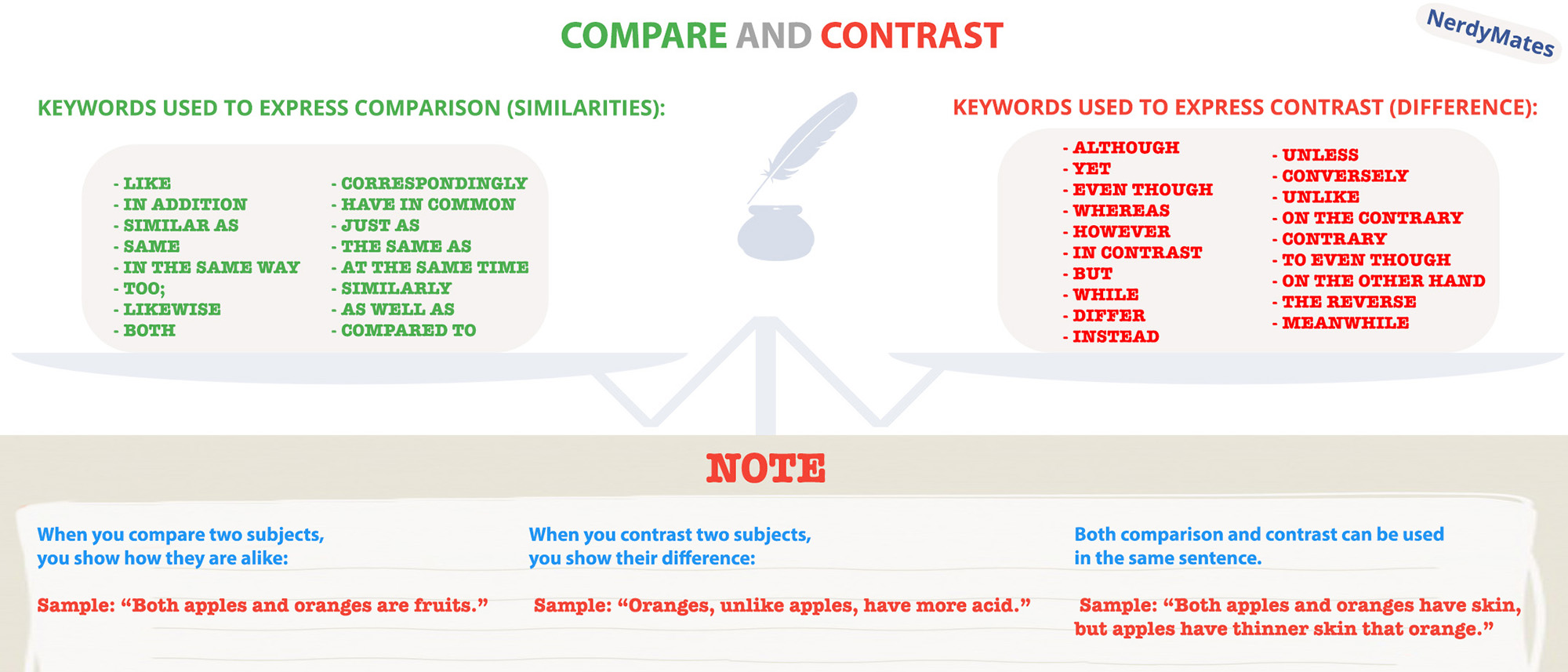
Generates a sample-to-sample comparison of SNPs from a vcf file - similarity-matrix.pl I am looking for a program that could help to compare two vcf files of two cultivars. I have three vcf files (vcf1, vcf2, vcf3) of SNPs of three different cultivars. The SNPs were called using GATK's UnifiedGenotyper. After stringent filtering with GATK's hard filters, I found approx. 6 millions SNPs in these three cultivars. Now I would like
I have two large multi-sample vcf files. The samples in both files are the same except the snp's were called using different programs. i want to see if they match. I am not sure if vcf-compare works. I gave it a shot and it returned all zeros. Is there any other way i can compare two multi-sample Welcome to VCFtools. VCFtools is a program package designed for working with VCF files, such as those generated by the 1000 Genomes Project. The aim of VCFtools is to provide easily accessible methods for working with complex genetic variation data in the form of VCF files.
I have 6 VCF files from different populations and I have sorted and compared them using VCF tools and tabix, but still the results are vast enough to … Sample VCF file for the test. sample.vcf This is a package of classes that are meant to provide an interface to access data of iCalendar and vCard files defined by the IMC (Internet Mail Consortium). The library can read and write files with the formats defined by IMC.
cd vcftools/ ./configure make make install This will compile both the PERL API and the C++ executable. By default, the compiled programs will now be in the vcftools/bin/ directory. Some common compiler errors may appear, such as not being able to locate Vcf… VCF to CSV - Convert VCF (Electronic business cards) file to CSV (Comma-Separated Values) file online for free - Convert document file online.
VCF to ODS - Convert VCF (Electronic business cards) file to ODS (OpenDocument Spreadsheet) file online for free - Convert document file online. VCF to CSV - Convert VCF (Electronic business cards) file to CSV (Comma-Separated Values) file online for free - Convert document file online.
SnpSift is a toolbox that allows you to filter and manipulate annotated files. For older version of this page, see: Manual page for SnpSift version 4.0 Once your genomic variants have been annotated, you need to filter them out in order to find the "interesting / relevant variants". A set of tools written in Perl and C++ for working with VCF files, such as those generated by the 1000 Genomes Project. - vcftools/vcftools
Table 1 summarizes the files that are generated by dbSNP, a brief overview of their content, frequency of updates and the location of the files for variations mapped to the most recent builds of GRCh37 and GRCh38. File names in Table 1. are … There is a program called vcf-compare in vcftools for comparing files. That's probably a good start, though you'll have to be more specific if that is not exactly what you want (vcf-stats may also be of interest).
I have 6 VCF files from different populations and I have sorted and compared them using VCF tools and tabix, but still the results are vast enough to … VCF to PDF - Convert VCF (Electronic business cards) file to PDF (Portable Document Format) file online for free - Convert PDF file online.
vcf-compare. Compares two multi entry vCard files and shows differences. Installation Instructions. No install needed. Just copy the file whereever you want. fill-rsIDs. Fill missing rsIDs. This script has been discontinued, please use vcf-annotate instead.. vcf-annotate. The script adds or removes filters and custom annotations to VCF files.
VCF to HTML - Convert VCF (Electronic business cards) file to HTML (Hypertext Markup Language) file online for free - Convert document file online. 29/08/2017В В· Thank you for the quick response!! The data is haploid, yes, because the organism (fungus) is haploid as well. What I want to achieve here is simply get the simplest statistics, e.g. the total number of SNPs per each sample (re-sequenced genome) in the VCF file.
Hello biostars! I was wondering, how to compare a few samples in my multisample vcf file? Basically, I would like to get a venn diagram, but vcf-compare is working only on multiple vcf files, bcftools stats and plot-vcfstats do not help either (they do not compare positions across the samples). VCF to CSV - Convert VCF (Electronic business cards) file to CSV (Comma-Separated Values) file online for free - Convert document file online.


